MINT - Metabolomics Integrator¶
MINT is a comprehensive toolkit for liquid chromatography-mass spectrometry (LC-MS) based metabolomics, providing both a powerful Python library and an intuitive web-based interface for extracting, visualizing, and analyzing targeted metabolomics data from complex biological samples.
Why MINT?¶
Metabolomics—the comprehensive study of small molecule metabolites in biological samples—plays a critical role in:
- Biomedical Research: Identifying biomarkers for disease diagnostics and therapeutic development
- Pathogen Detection: Distinguishing bacterial strains (e.g., methicillin-resistant Staphylococcus aureus [MRSA])
- Drug Discovery: Understanding metabolic pathways and drug effects
- Environmental Science: Tracking metabolic responses to environmental changes
MINT streamlines the LC-MS data processing workflow with powerful features for targeted metabolite quantification, quality assessment, and statistical analysis. It is particularly well-suited for handling large amounts of data (10,000+ files).
Quick Links¶
- Python Library Documentation - Core ms-mint library for scripts and notebooks
- Web Application Documentation - Browser-based MINT interface
- Demo Data - Download test data with sample LC-MS files and target lists
Resources¶
- Lewis Research Group Software - Explore additional metabolomics and computational biology tools
- GitHub: ms-mint - Core Python library
- GitHub: ms-mint-app - Web application
- Plugin Template - Extend MINT functionality
Understanding LC-MS Metabolomics¶
The Challenge¶
Biological samples (e.g., blood, tissue, bacterial cultures) contain thousands of metabolites—sugars, amino acids, nucleotides, lipids, and more. Mass spectrometry can measure these compounds with high sensitivity and accuracy, but many metabolites share identical or very similar masses, making them indistinguishable by mass alone.
The Solution: Liquid Chromatography¶
To separate metabolites before mass spectrometry analysis, liquid chromatography (LC) is used. As the sample flows through a chromatographic column, metabolites interact differently with the column material based on their chemical properties. This causes metabolites to elute (exit the column) at different retention times, spreading them out over time so they can be measured individually by the mass spectrometer.
LC-MS Data Structure¶
The mass spectrometer continuously measures ion intensities across a range of mass-to-charge (m/z) values as metabolites elute from the column. This produces a three-dimensional dataset: retention time, m/z, and intensity.
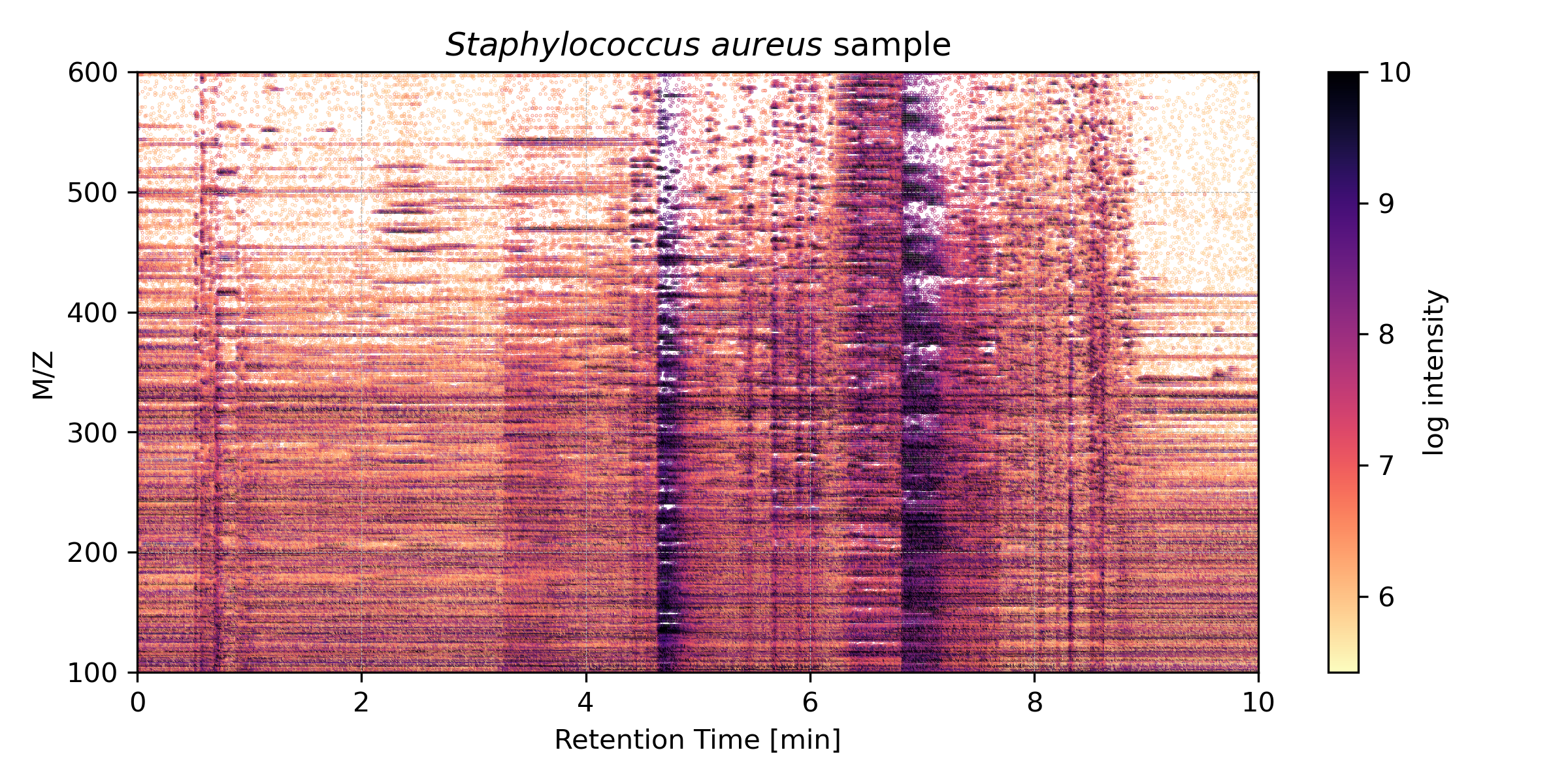 Figure 1: 2D heatmap of LC-MS data from _S. aureus showing ion intensities over 10 minutes for m/z 100–600. Brighter colors indicate higher ion abundance._
Figure 1: 2D heatmap of LC-MS data from _S. aureus showing ion intensities over 10 minutes for m/z 100–600. Brighter colors indicate higher ion abundance._
Zooming into a narrow m/z range reveals individual metabolite peaks. For example, here is the extracted ion chromatogram for succinate (succinic acid):
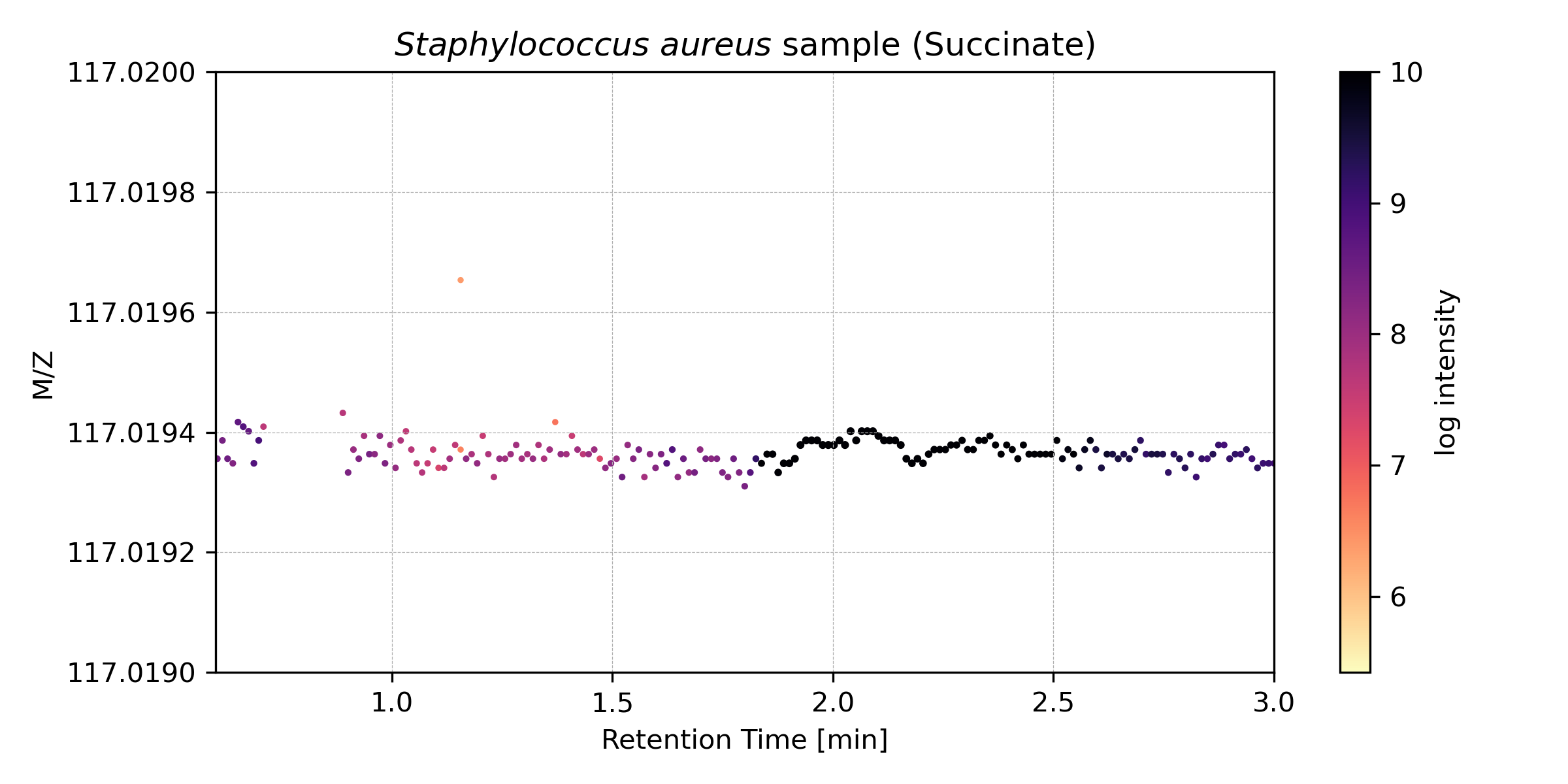 Figure 2: Zoomed view showing the chromatographic peak for succinate. The sharp peak indicates high signal and good chromatographic separation.
Figure 2: Zoomed view showing the chromatographic peak for succinate. The sharp peak indicates high signal and good chromatographic separation.
This demonstrates the precision of LC-MS data—mass measurements are accurate to fractions of a Dalton (for comparison, an electron has m/z = 5.489×10⁻⁴).
How MINT Processes LC-MS Data¶
Data Conversion¶
Raw LC-MS data is typically stored in vendor-specific formats (e.g., Thermo .raw, Agilent .d). MINT requires data to be converted to open formats: - mzML (preferred) - mzXML
Most vendor software provides conversion tools, or you can use open-source converters like MSConvert.
Targeted Peak Extraction¶
Rather than analyzing the entire LC-MS dataset, MINT uses a targeted approach:
- Define targets: Specify metabolites by their expected m/z and retention time (RT)
- Extract peaks: MINT integrates ion intensities within defined m/z and RT windows
- Quantify: Peak areas or heights are calculated, proportional to metabolite abundance
Important Note on Quantification: - Peak intensities reflect relative abundance within the same metabolite across samples - Intensities cannot be compared between different metabolites (due to varying ionization efficiencies) - For absolute quantification, calibration curves with known standards are required
Data Structuring and Analysis¶
MINT transforms raw 3D LC-MS data into a structured table where: - Rows = samples - Columns = metabolites - Values = peak areas/heights
This structured format enables downstream statistical analyses: - Normalization and scaling - Principal Component Analysis (PCA) - Hierarchical clustering - Statistical testing
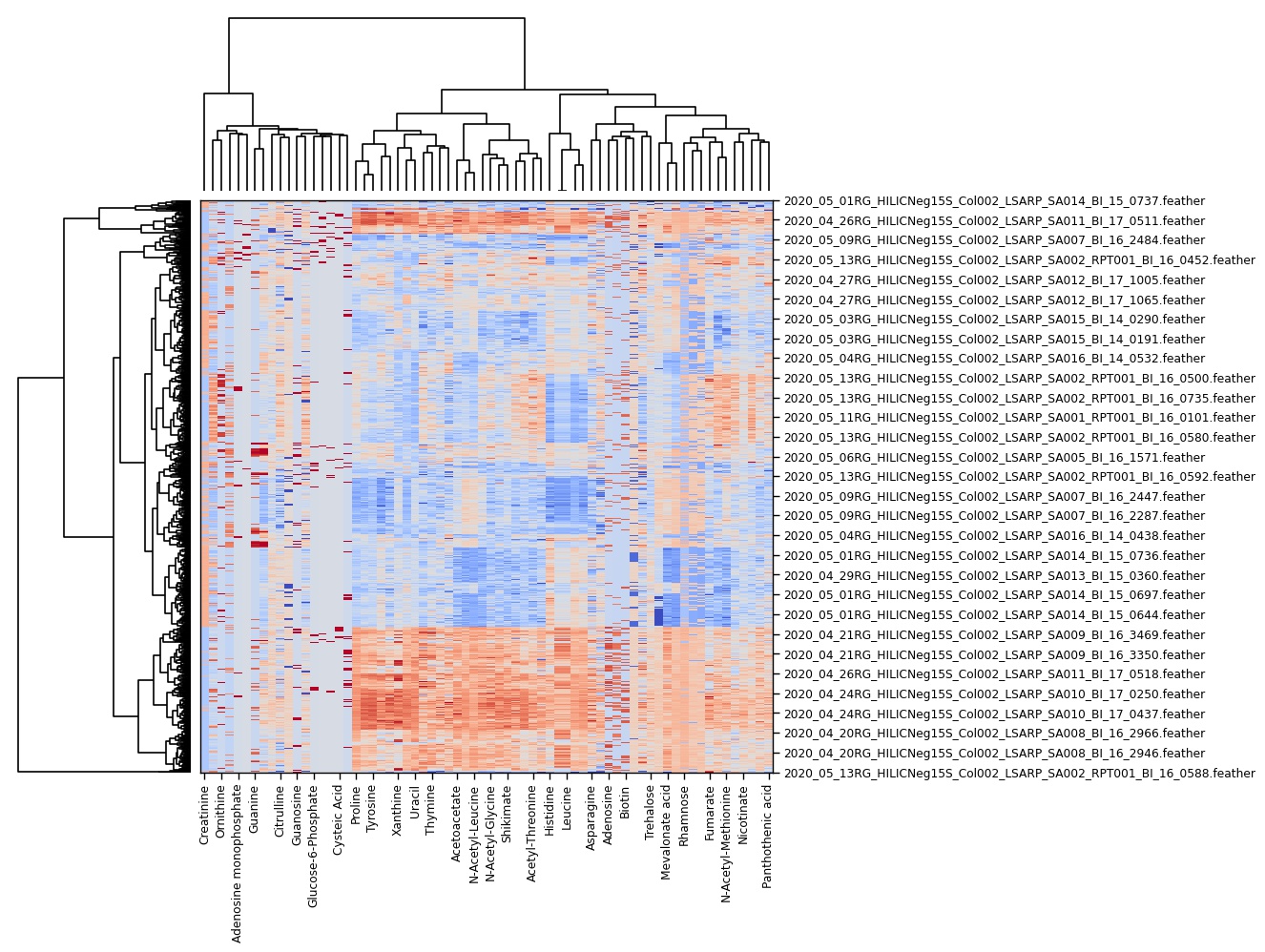 Figure 3: Hierarchical clustering heatmap showing metabolic profiles of 12 samples from three bacterial species: _E. coli (EC), S. aureus (SA), and C. albicans (CA). Samples cluster by organism, demonstrating species-specific metabolic signatures._
Figure 3: Hierarchical clustering heatmap showing metabolic profiles of 12 samples from three bacterial species: _E. coli (EC), S. aureus (SA), and C. albicans (CA). Samples cluster by organism, demonstrating species-specific metabolic signatures._
Citation¶
When using MINT in your research, please cite:
- ms-mint library: DOI: 10.5281/zenodo.12733875
- ms-mint-app: DOI: 10.5281/zenodo.13121148
Support¶
Contributing¶
MINT is an open-source project. Contributions are welcome!
- Report issues on GitHub
- Submit pull requests
- Share improvements and extensions
Disclaimer¶
MINT is provided as-is. Always validate results and consult domain experts.